Funding
NEUROMODEL project
I am the happy beneficiary of an EU grant Marie Sklodowska-Curie Individual Fellowship (MSCA-IF) for the period June 2017 - June 2019. The project aims to meet the need in neuroscience for flexible statistical tools able to jointly analyze clinical, genetic, psychological and imaging data.
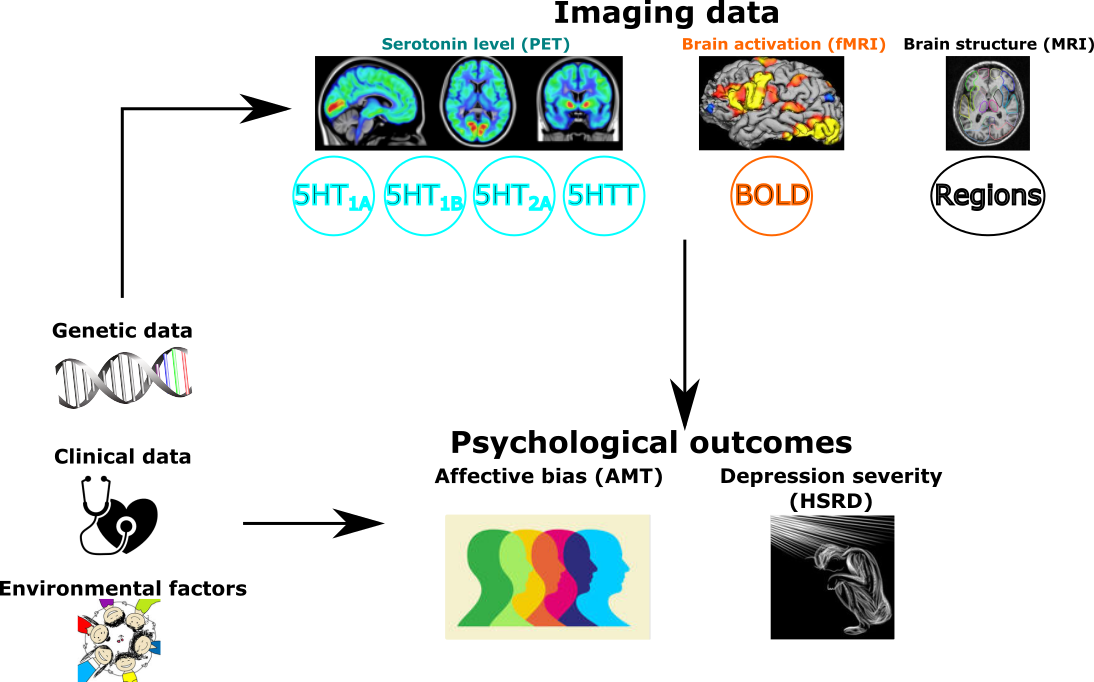 Variables involved in an integrative model of depression.
Variables involved in an integrative model of depression.
LVMs is a tool of choice for studying systems of variables and in particular when considering noisy or indirect measurements of the quantity of interest. This is typically the case when studying depression: we cannot directly measure it but we can observe its effect on memory, reaction time, … . LVMs define unobserved variables such as depression as latent variables and attempt to identify them using the observed data and the modeling assumptions.
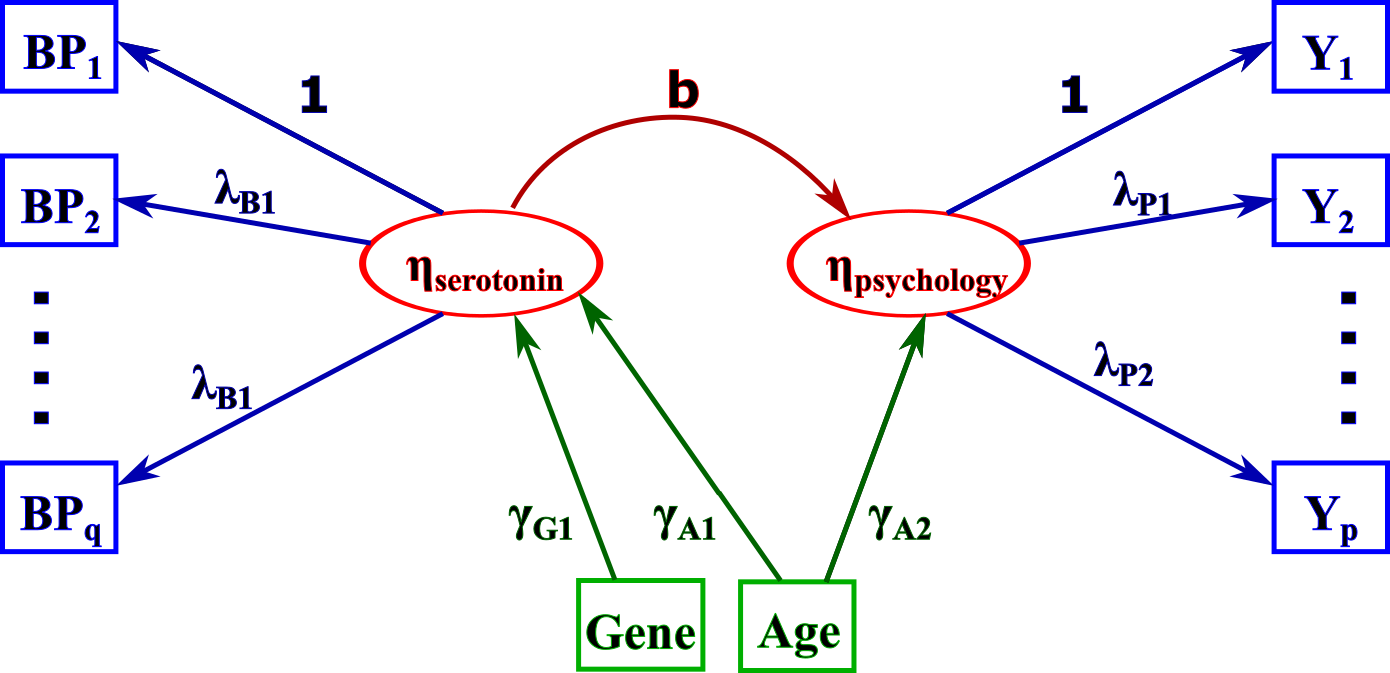 Example of LVM: the square boxes represent observed variables while the round boxes contain the latent variables. BP denotes binding potential and the Y some psychological measurements (e.g. memory). This LVM can be used to study the impact of serotonin on the mood when only indirect measurements of both quantities are available.
Example of LVM: the square boxes represent observed variables while the round boxes contain the latent variables. BP denotes binding potential and the Y some psychological measurements (e.g. memory). This LVM can be used to study the impact of serotonin on the mood when only indirect measurements of both quantities are available.
The project aims at adapting LVMs to the context of neuroscience. This is a particular challenging field because studies have often a very limited number of participants (typically less than 100) and involve many variables with intricate correlation structures. The project is divided in three work packages (WPs), the first two focus on statistical/software developments and the third on dissemination and application in neuroscience.
Work package 1: regularized LVMs
As often in statistics, LVMs provide correct estimates when they are correctly specified, i.e. we correctly model the relationship between variables. However a correct specification usually requires more background knowledge than available. A data-driven procedure is therefore often used to check the validity of the proposed model. The traditional method, forward stepwise search, is not completely satisfying:
- it is a greedy approach (only explores specific subsets of the space possible models so it may not find the optimal solution)
- it becomes intractable when a large number of alternative models can be considered.
Regularization is an alternative technic that has been successfully applied in univariate models. WP1 is about the development of regularized LVMs. This has been done through the development of the lavaPenalty package for the R software. It enables to fit LVMs with lasso/ridge/nuclear norm penalties and provide some tools for estimating the regularization path. Nevertheless to be applicable in clinical studies it currently lacks a framework for performing statistical inference. This will be investigated in WP2.
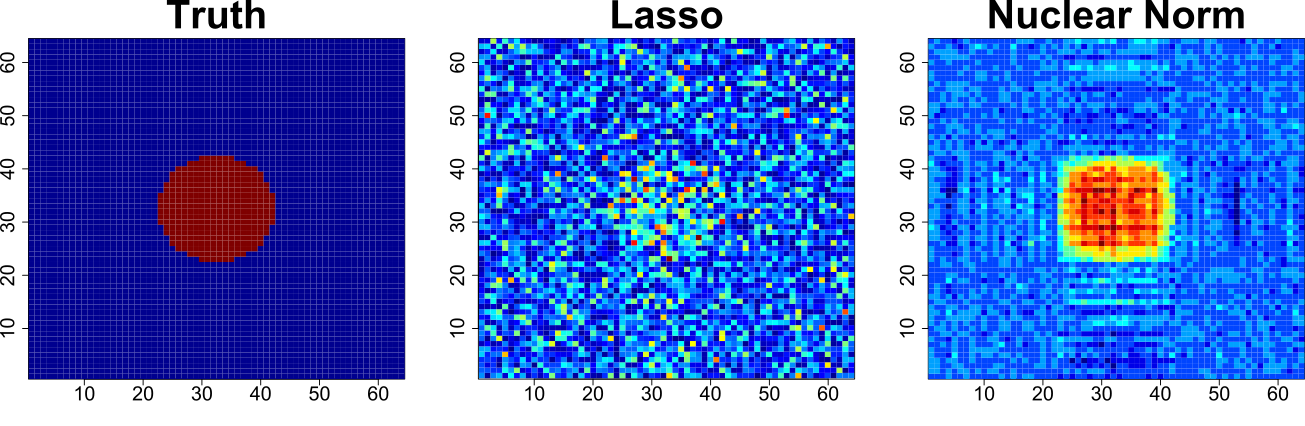 Comparison
between lasso and nuclear norm regularization for fitting a geometric
shape.
Comparison
between lasso and nuclear norm regularization for fitting a geometric
shape.
Work package 2: Inference in LVMs
The original aim of this work package was to better understand the statistical properties of estimators in regularized LVMs. But, to begin with, I first studied the behavior of statistical tests used with LVM traditionally in neuroscience studies. These studies are characterized by small samples and multiple hypotheses to test, which make the traditional asymptotic results from maximum likelihood theory difficult to apply.
In a first article (currently in revision in JRSS-C) I developed a correction for the bias of the maximum likelihood estimator of the variance parameters, and derived the degrees of freedom of the corresponding variance parameters. Then, as in univariate linear models or when using the Kenward and Roger correction in mixed model, I proposed to model the Wald test statistic of LVMs as Student’s t-distributed distributed (instead of normally distributed). This leads to an improved control of the type 1 error.
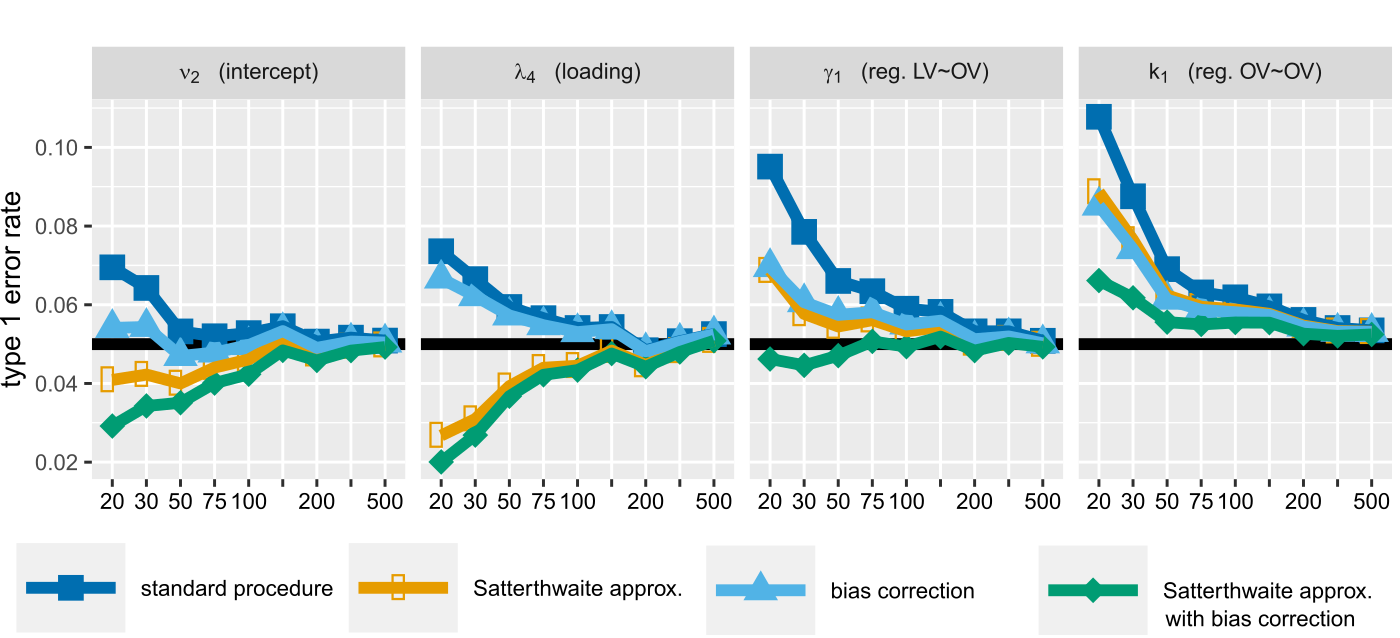 Control of the
type 1 error in a latent factor model using no correction (dark blue)
or the proposed correction (green).
Control of the
type 1 error in a latent factor model using no correction (dark blue)
or the proposed correction (green).
In a second article (to be submitted to Psychometrika) I addressed the problem of controlling the type 1 error when performing multiple comparisons. One solution is to use a Bonferroni correction but this solution is known to be too conservative. This is unsatisfactory in clinical studies for ethical and economical reasons. Fortunately, correlation can be accounted for using max-test procedures: while still controlling the type 1 error max-test procedure are more powerful than Bonferroni procedure. I therefore integrate max-test procedures in the LVM framework for both Wald tests and score tests.
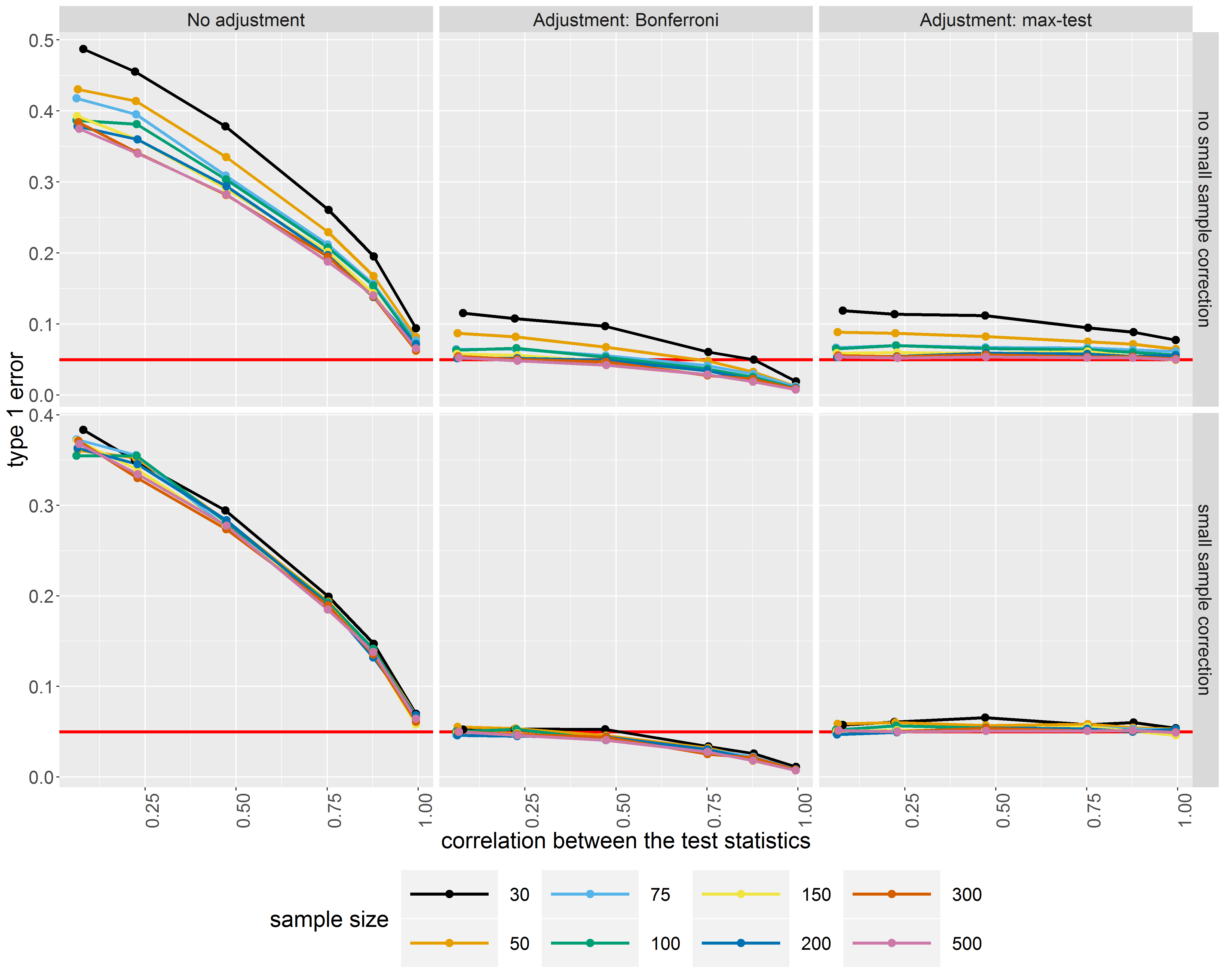 Control of
the type 1 error when performing multiple testing using or not a
correction for multiple testing (columns) and for the small sample
bias of maximum likelihood estimators (rows).
Control of
the type 1 error when performing multiple testing using or not a
correction for multiple testing (columns) and for the small sample
bias of maximum likelihood estimators (rows).
These two development are available in the package lavaSearch2. I are now focusing my work on inference after regularization and selection.
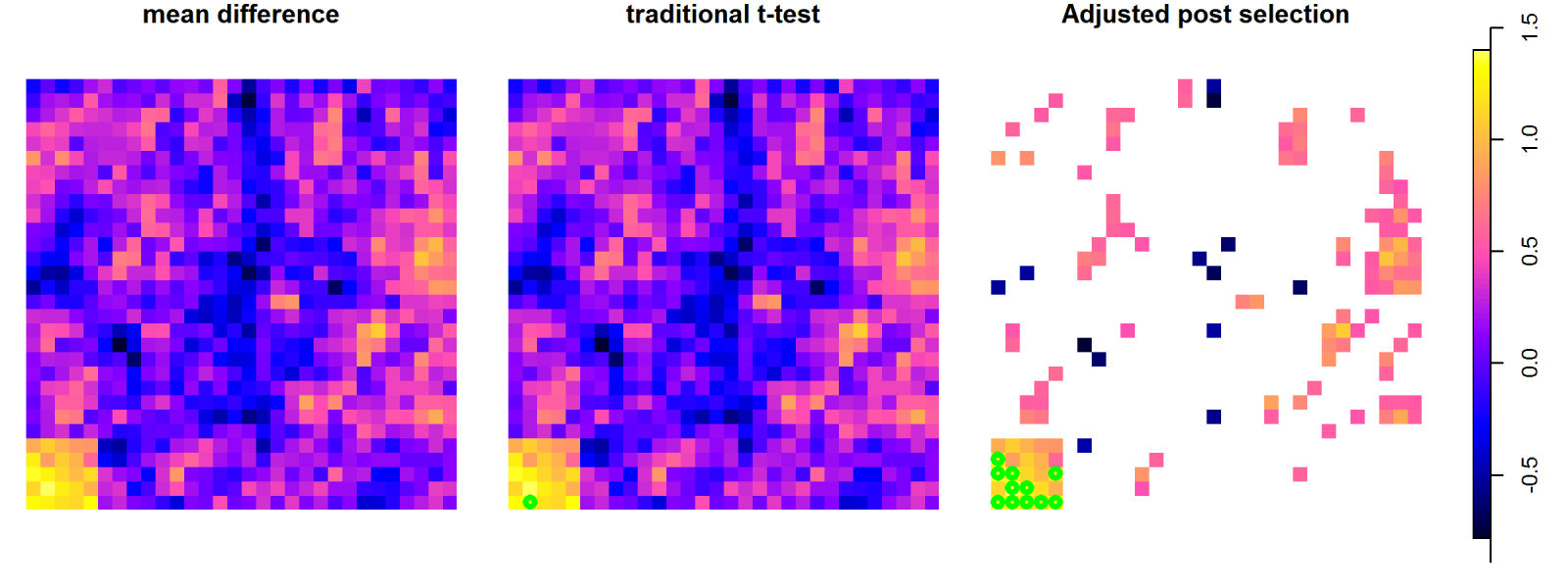 Comparison between the signal in two groups at the voxel level, using
a t-test at each voxel or only at voxels with a signal greater than a
pre-defined value. The green dots refer to the voxels where a significant
difference is identified.
Comparison between the signal in two groups at the voxel level, using
a t-test at each voxel or only at voxels with a signal greater than a
pre-defined value. The green dots refer to the voxels where a significant
difference is identified.
Work package 3: Dissemination and application in neuroscience
The last WP aimed at disseminating the developped method in neurosience, in particular to neuroscientists at the Neurobiology Reasearch Unit. There, I developped several collaborations:
- with Sebastian Ebert who is interested in the consequences of concussion in term of neuroinflammation. For this project, I use both the small sample correction and the adjustment for multiple comparisons developped in WP2 to perform the statistical analysis. The overall conclusion of the study is that concussion was associated with elevated 123I-CLINDE binding to TSPO, an indicator of brain inflammatory response. This work has been published in the european journal of neurology (https://doi.org/10.1111/ene.13971).
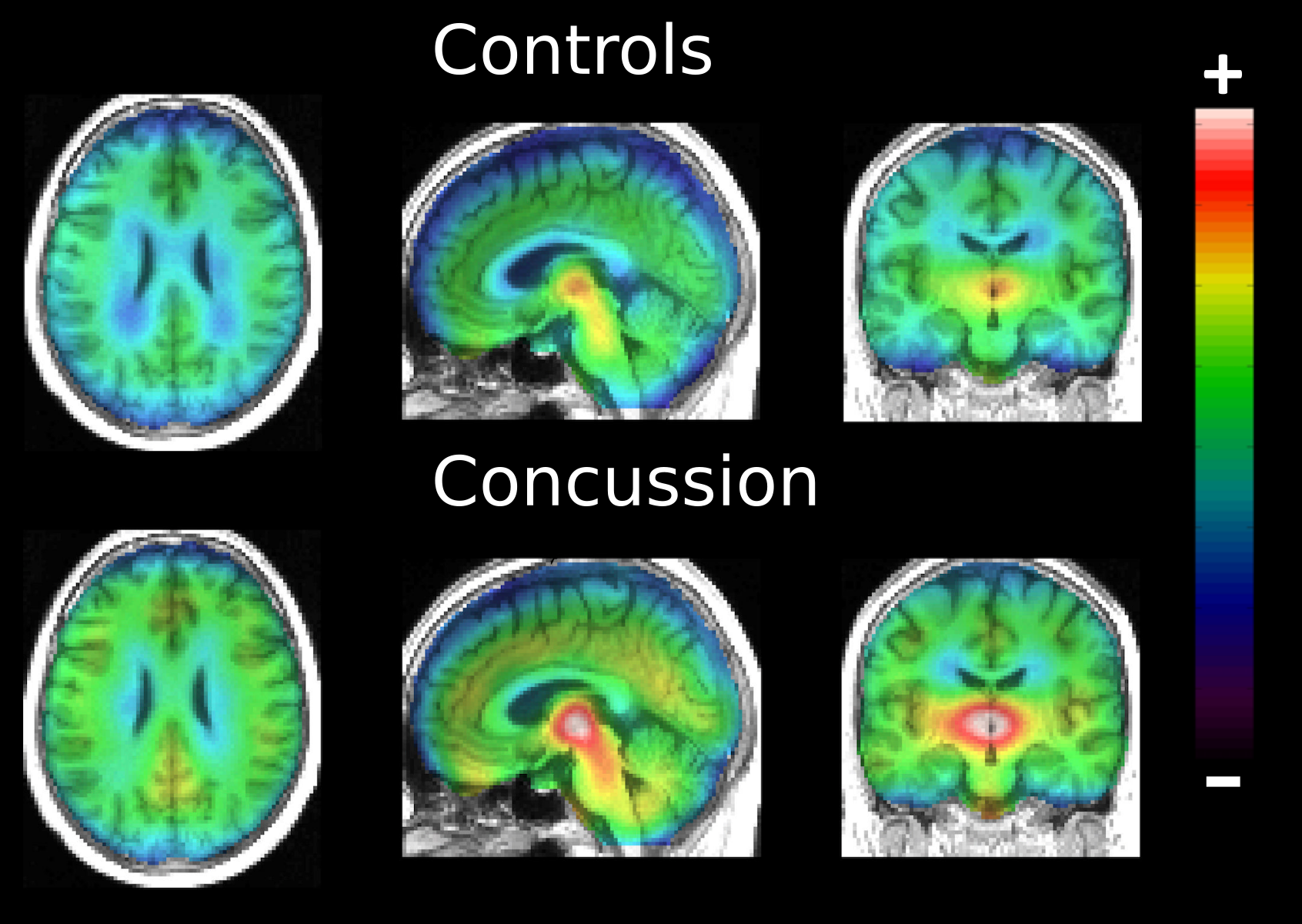 Normalized 123I-CLINDE across groups.
Normalized 123I-CLINDE across groups.
- with Martin Nørgaard who works on integrating the uncertainty related to the pre-processing of the data in medical data analysis. Indeed, raw data are typically transformed to remove measurement artefacts, increase signal to noise ratio, or normalize data across individuals. However there are several method available which may make the results depend on the investigator choice of pre-processing pipeline. Taking the example of a classifier trained on imaging data, we show that the choice of the pre-processing method have a critical impact that should be accounted for when assessing its performance. This is a slight modification of the max-test procedure used in WP2; here we use permutations to obtain a non-parametric procedure. This article has been accepted at the MICCAI conference 2019.
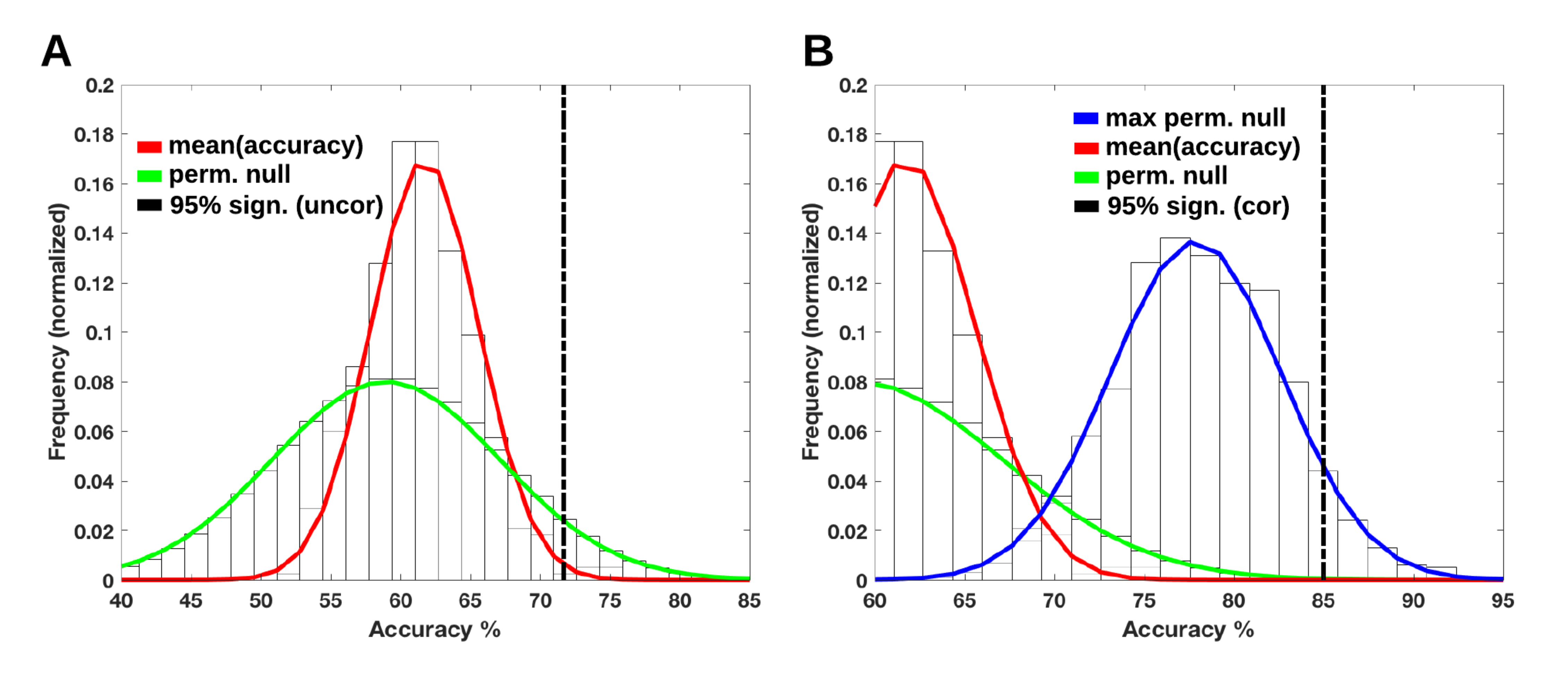 Classification accuracies for different pre-processing
pipelines. Accuracies are estimated from a Monte-Carlo experiment;
only the average accuracy is reported (red curve). The green curve
represent the distribution of the accuracy under the null hypothesis
for a single pipepline while the blue curve uses the best pipeline.
Classification accuracies for different pre-processing
pipelines. Accuracies are estimated from a Monte-Carlo experiment;
only the average accuracy is reported (red curve). The green curve
represent the distribution of the accuracy under the null hypothesis
for a single pipepline while the blue curve uses the best pipeline.
- with Vincent Beliveau who works on identifying brain regions specific to the serotonin system. I assisted him in developping a clustering algorithm flexible enough to capture the spatial patterns of the serotonin system, and finding the appropriate metrics to assess the quality of the segmentation. The corresponding article is currently is revision in Neuroimage.
Two projects with Liv V. H. Bruel on their way: one where we compare the structure of memory between “normal” individuals and aggressive people and another where we identified stable concepts between two versions of a questionnaire measuring short, intermediate, and long-term memory.
Thanks to a collaboration with research from the section of biostatistics from the University of Copenhagen, I have also been working on deriving estimators robust to model misspecification. One result of this work is ate function from the riskRegression that can be use to estimate averate treatment effect, even in the presence of missing values. The corresponding manuscript is currently in revision in the Biometrical journal.
 Comparison of various estimators of the average treatment
effect. Three statistical models can be specified: one for the risk of
getting the disease, one for the treatment allocation, and one for the
probability of dropping out from the study. The AIPTW.AIPCW estimator
is robust (i.e. unbiased) to the misspecification of one of the statistical
models.
Comparison of various estimators of the average treatment
effect. Three statistical models can be specified: one for the risk of
getting the disease, one for the treatment allocation, and one for the
probability of dropping out from the study. The AIPTW.AIPCW estimator
is robust (i.e. unbiased) to the misspecification of one of the statistical
models.
This approach, and in particular the ate function, is being used by our clinical collaborators to study the effect of beta blockers vs. alternative antihypertensives on the risk of Alzheimer disease and to study risk factor of cardio-vascular diseases based data from on the danish national patient registry.
Training and dissemination
Thanks to the grant I have been able to attend the following workshops:
- Advanced Topics in Machine Learning 2016, DTU compute (Lyngby, Denmark; August 28th - September 1st, 2018).
- Survival Analysis for Junior Researchers (Copenhagen, Denmark; April 24th - 26th, 2019).
- Mediation analysis, pre-conference course of the Nordic-Baltic Biometric (Vilnius, Lithuania, 2 June 2019).
- Analysis of interval-censored data, pre-conference course of the Annual Conference of the International Society for Clinical Biostatistics (Leven, Belgium; July 14 2019).
and to present my work at international conferences:
- 40th Annual Conference of the International Society for Clinical Biostatistics, 14-18 July 2019, Leuven, Belgium: Multiple testing in latent variable models: Dunnett adjustment and small sample correction (contributing speaker)
- 7th Nordic-Baltic Biometric Conference, 3-5 June 2019, Vilnius, Lithuania: Region-based and voxel-wise analysis of medical images using latent variables (invited speaker)